solo para uso en investigación
Pioglitazone (U-72107) Agonista de PPARγ
Cat. No.S2590
Estructura química
Peso molecular: 356.44
Control de calidad
| Dianas relacionadas | Dehydrogenase HSP Transferase P450 (e.g. CYP17) PDE phosphatase Vitamin Carbohydrate Metabolism Mitochondrial Metabolism Drug Metabolite |
|---|---|
| Otros PPAR Inhibidores | T0070907 GW9662 GW6471 WY-14643 (Pirinixic Acid) GSK3787 GW0742 AZ6102 Harmine Astaxanthin Eupatilin |
Cultivo celular, tratamiento y concentración de trabajo
| Líneas celulares | Tipo de ensayo | Concentración | Tiempo de incubación | Formulación | Descripción de la actividad | PMID |
|---|---|---|---|---|---|---|
| CV-1 | Function assay | In vitro transcriptional activation of Peroxisome proliferator activated receptor gamma (PPAR) expressed in CV-1 cells, EC50=0.69μM | 8576907 | |||
| 3T3-L1 | Function assay | Effective concentration for 50% enhancement of insulin-induced triglyceride accumulation in 3T3-L1 cells, EC50=0.16μM | 9599241 | |||
| CV-1 | Function assay | Activation of peroxisome proliferator activated receptor gamma measured by induction of 50% of maximum alkaline phosphatase activity, transfection assay in CV-1 cells, EC50=0.58884μM | 9836620 | |||
| CV-1 | Function assay | Maximal reporter activity against human Peroxisome proliferator activated receptor gamma Gal4 chimeric in transiently transfected CV-1 cells by functional assay., EC50=0.58μM | 11720854 | |||
| HEK293 | Function assay | Agonist activity at PPARgamma expressed in HEK293 cells assessed as induction of receptor interaction with steroid receptor coactivator-1 by EYFP based reporter gene assay | 16680159 | |||
| HEK293 | Function assay | Agonist activity at PPARgamma expressed in HEK293 cells assessed as induction of receptor interaction with retinoid X-receptor alpha by EYFP based reporter gene assay | 16680159 | |||
| CV1 | Function assay | Transactivation of PPARgamma in CV1 cells, EC50=0.55μM | 16821769 | |||
| Cos7 | Function assay | 14 hrs | Transactivation of human PPARgamma LBD expressed in african green monkey Cos7 cells co-transfected with fused GAL4-DBD after 14 hrs by Dual-Glo Luciferase reporter gene assay, EC50=0.3μM | 20307981 | ||
| 3T3L1 | Function assay | 100 umol/L | Increase in PPARgamma mRNA levels in mouse 3T3L1 cells at 100 umol/L by RT-PCR | 20392645 | ||
| CHO | Function assay | Activation of Gal4-tagged human PPARgamma expressed in CHO cells by luciferase reporter gene assay, EC50=0.14μM | 20656494 | |||
| HEK293T | Function assay | 1 uM | 24 hrs | Partial agonist activity at human PPARgamma-LBD expressed in HEK293T cells assessed as induction of receptor transactivation at 1 uM after 24 hrs by luciferase reporter gene assay | 21030263 | |
| COS-1 | Function assay | Agonist activity at human PPARgamma ligand binding domain expressed in COS-1 cells co-transfected with Gal4 by luciferase reporter gene assay, EC50=0.39μM | 21130649 | |||
| CHO | Function assay | 24 hrs | Partial agonist activity at human PPARgamma expressed in CHO cells co-transfected with Gal4-responsive luciferase reporter plasmid after 24 hrs by transactivation assay, EC50=0.39μM | 21377875 | ||
| COS7 | Function assay | Modulation of human PPARgamma-LBD expressed in african green monkey COS7 cells co-transfected with Gal4 assessed as activation of transactivation activity by luciferase assay, EC50=0.3μM | 21873070 | |||
| Sf21 | Function assay | Inhibition of human BSEP expressed in plasma membrane vesicles of Sf21 cells assessed as inhibition of ATP-dependent [3H]taurocholate uptake, IC50=0.3μM | 21965623 | |||
| Sf21 | Function assay | Inhibition of Sprague-Dawley rat Bsep expressed in plasma membrane vesicles of Sf21 cells assessed as inhibition of ATP-dependent [3H]taurocholate uptake, IC50=2μM | 21965623 | |||
| HepG2 | Function assay | 20 hrs | Agonist activity at GAL4-tagged human PPARgamma ligand binding domain expressed in HepG2 cells assessed as transactivation after 20 hrs by beta-galactosidase reporter gene assay, EC50=0.57μM | 22341573 | ||
| HEK293 | Function assay | 18 hrs | Transactivation of human GAL4-fused PPARgamma ligand binding domain transfected in HEK293 cells after 18 hrs by dual luciferase reporter gene assay, EC50=0.8μM | 23102891 | ||
| COS7 | Function assay | Transactivation of GAL4-fused human PPARgamma ligand binding domain transfected in african green monkey COS7 cells by luciferase reporter gene assay, EC50=0.2μM | 23130964 | |||
| THP1 | Antiinflammatory assay | 1 hr | Antiinflammatory activity in human THP1 cells assessed as inhibition of PMA-induced MCP-1 secretion preincubated for 1 hr prior PMA-challenge measured after 48 hrs by ELISA, IC50=18.84μM | 23811092 | ||
| THP1 | Antiinflammatory assay | 2 hrs | Antiinflammatory activity in human THP1 cells assessed as inhibition of PMA-induced MCP-1 secretion incubated for 2 hrs prior to PMA challenge measured after 72 hrs by ELISA, IC50=18.6μM | 24531227 | ||
| HEK293 | Function assay | 10 uM | 24 hrs | Transactivation of PPAR-gamma (unknown origin) expressed in HEK293 cells at 10 uM after 24 hrs by luciferase reporter gene assay | 24890090 | |
| HEK293 | Function assay | 18 hrs | Agonist activity at human PPARgamma expressed in HEK293 cells incubated for 18 hrs by luciferase reporter gene assay, EC50=0.21μM | 25333853 | ||
| COS7 | Function assay | Transactivation of human PPARgamma expressed in African green monkey COS7 cells incubated overnight by dual-glo luciferase reporter gene assay, EC50=0.2μM | 26595749 | |||
| THP1 | Function assay | 10 uM | 24 hrs | Upregulation of ABCA1 mRNA expression in human THP1 cells at 10 uM after 24 hrs by qPCR method relative to control | 27220065 | |
| THP1 | Function assay | 10 uM | 24 hrs | Upregulation of ABCG1 mRNA expression in human THP1 cells at 10 uM after 24 hrs by qPCR method relative to control | 27220065 | |
| COS7 | Function assay | 42 hrs | Transactivation of GAL4-fused human PPARgamma ligand binding domain expressed in African green monkey COS7 cells after 42 hrs by dual luciferase reporter gene assay, EC50=0.5μM | 27560282 | ||
| COS7 | Function assay | 42 hrs | Transactivation at Gal4 fused PPARgamma LBD (unknown origin) expressed in African green monkey COS7 cells after 42 hrs by luciferase assay, EC50=0.32μM | 27569195 | ||
| COS7 | Function assay | 42 hrs | Transactivation of Gal4 fused human PPARgamma LBD expressed in African green monkey COS7 cells after 42 hrs by dual luciferase reporter gene assay, EC50=0.38μM | 27918994 | ||
| PC3 | Function assay | 50 uM | 24 hrs | Increase in p21(WAF1) protein expression in human PC3 cells at 50 uM incubated for 24 hrs by Western blot analysis | 28395220 | |
| MDA-MB-231 | Function assay | 50 uM | 24 hrs | Increase in p21(WAF1) protein expression in human MDA-MB-231 cells at 50 uM incubated for 24 hrs by Western blot analysis | 28395220 | |
| HEK293 | Function assay | 24 hrs | Transactivation activity at Gal4 fused full length human PPARgamma LBD expressed in HEK293 cells after 24 hrs by luciferase reporter gene assay, EC50=1.1μM | 28465099 | ||
| HEK293 | Function assay | 4 hrs | Transrepression activity at human PPARgamma expressed in HEK293 cells assessed as inhibition of TNFalpha induced NF-kappaB promoter activity pretreated for 4 hrs followed by TNFalpha stimulation after 3 hrs by luciferase reporter gene assay, IC50=22μM | 28465099 | ||
| SCC15 | Cytotoxicity assay | 50 uM | 24 hrs | Cytotoxicity against human SCC15 cells transfected with PPARalpha siRNA assessed as reduction in cell viability at 50 uM after 24 hrs by resazurin reduction assay | 29031063 | |
| SCC15 | Cytotoxicity assay | 50 uM | 24 hrs | Cytotoxicity against human SCC15 cells transfected with PPARbeta siRNA assessed as reduction in cell viability at 50 uM after 24 hrs by resazurin reduction assay | 29031063 | |
| HepG2 | Function assay | 24 hrs | Agonist activity at PPARgamma in human HepG2 cells assessed as activation of PPRE incubated for 24 hrs by dual luciferase reporter gene assay, EC50=0.24μM | 30001846 | ||
| HEK293BENA | Function assay | 24 hrs | Transactivation of GAL4-fused human PPARgamma transfected in HEK293BENA cells after 24 hrs by steady glo-luciferase reporter gene assay, EC50=2.053μM | 30351933 | ||
| HEK293 | Function assay | 18 hrs | Transactivation of human full length PPARgamma expressed in HEK293 cells after 18 hrs by luciferase reporter gene based luminescence assay, EC50=0.1μM | 30362739 | ||
| 293H | Function assay | 16 hrs | Transactivation of GAL4-DBD fused human PPARgamma ligand binding domain expressed in UAS-bla HEL 293H cells preincubated for 16 hrs followed by FRET substrate addition and measured after 2 hrs by TR-FRET assay, EC50=0.098μM | 30429097 | ||
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in AP1 mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in CD36 mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in Glut4 mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in adiponectin mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| stem cells | Function assay | 5 days | Induction of adipogenesis in human bone marrow-derived mesenchymal stem cells assessed as increase in adiponectin production measured on day 5 in presence of IDX by ELISA, EC50=0.35μM | 30672698 | ||
| HEK293T | Function assay | 18 hrs | Transactivation of full length human PPARgamma2 expressed in HEK293T cells co-expressing PPRE after 18 hrs by luciferase reporter gene assay, EC50=0.25μM | 30676741 | ||
| HEK293T | Function assay | 18 hrs | Transactivation of chimeric Gal4 yeast DBD fused-PPARgamma LBD (unknown origin) expressed in HEK293T cells co-expressing PPRE after 18 hrs by luciferase reporter gene assay, EC50=0.35μM | 30676741 | ||
| HepG2 | Function assay | 30 uM | Binding affinity to NAF1 (unknown origin) expressed in human HepG2 cells assessed as inhibition of mitochondrial respiration at 30 uM | 30770154 | ||
| HepG2 | Function assay | 30 uM | Binding affinity to NAF1 in human HepG2 cells assessed as inhibition of mitochondrial respiration at 30 uM | 30770154 | ||
| COS7 | Function assay | 39 hrs | Transactivation of Gal4-fused human PPARgamma transfected in COS7 cells co-transfected with pGAL5-TK-pGL3 and pRennilla-CMV incubated for 39 hrs by dual luciferase reporter assay, EC50=1.4μM | 31648125 | ||
| K562-IMA[r] | Function assay | 10 uM | 72 hrs | Induction of sensitization to imatinib-induced cytotoxicity in imatinib-resistant human K562-IMA[r] cells assessed as induction of cell death at 10 uM after 72 hrs in presence of 1 uM imatinib by propidium iodide/Triton-X100 dye based FACS analysis | 31648125 | |
| Haga clic para ver más datos experimentales de líneas celulares | ||||||
Información química, almacenamiento y estabilidad
| Peso molecular | 356.44 | Fórmula | C19H20N2O3S |
Almacenamiento (Desde la fecha de recepción) | |
|---|---|---|---|---|---|
| Nº CAS | 111025-46-8 | Descargar SDF | Almacenamiento de soluciones madre |
|
|
| Sinónimos | AD-4833, U 72107 | Smiles | CCC1=CN=C(C=C1)CCOC2=CC=C(C=C2)CC3C(=O)NC(=O)S3 | ||
Solubilidad
|
In vitro |
DMSO
: 20 mg/mL
(56.11 mM)
Water : Insoluble Ethanol : Insoluble |
Calculadora de Molaridad
|
In vivo |
|||||
Calculadora de formulación in vivo (Solución clara)
Paso 1: Introduzca la información a continuación (Recomendado: Un animal adicional para tener en cuenta la pérdida durante el experimento)
Paso 2: Introduzca la formulación in vivo (Esto es solo la calculadora, no la formulación. Por favor, contáctenos primero si no hay una formulación in vivo en la sección de Solubilidad.)
Resultados del cálculo:
Concentración de trabajo: mg/ml;
Método para preparar el líquido maestro de DMSO: mg fármaco predissuelto en μL DMSO ( Concentración del líquido maestro mg/mL, Por favor, contáctenos primero si la concentración excede la solubilidad del DMSO del lote del fármaco. )
Método para preparar la formulación in vivo: Tomar μL DMSO líquido maestro, luego añadirμL PEG300, mezclar y clarificar, luego añadirμL Tween 80, mezclar y clarificar, luego añadir μL ddH2O, mezclar y clarificar.
Método para preparar la formulación in vivo: Tomar μL DMSO líquido maestro, luego añadir μL Aceite de maíz, mezclar y clarificar.
Nota: 1. Por favor, asegúrese de que el líquido esté claro antes de añadir el siguiente disolvente.
2. Asegúrese de añadir el (los) disolvente(s) en orden. Debe asegurarse de que la solución obtenida, en la adición anterior, sea una solución clara antes de proceder a añadir el siguiente disolvente. Se pueden utilizar métodos físicos como el vórtice, el ultrasonido o el baño de agua caliente para ayudar a la disolución.
Mecanismo de acción
| Targets/IC50/Ki |
PPARγ
|
|---|---|
| In vitro |
La Pioglitazone es metabolizada principalmente por CYP2C8 y en menor medida por CYP3A4 in vitro. |
| In vivo |
La Pioglitazone atenúa significativamente la dilatación y disfunción de la cavidad ventricular izquierda (VI) mediante ecocardiografía, así como la presión telediastólica del VI en ratones con infarto de miocardio anterior extenso. Este compuesto normaliza parcialmente los dP/dt(max) y dP/dt(min) del VI, índices de la función contráctil del VI, que están significativamente reducidos en ratones con IM. Este compuesto da como resultado una activación reducida de la microglía, una inducción reducida de células positivas para iNOS y menos células positivas para la proteína glial fibrilar ácida tanto en el estriado como en la sustancia negra pars compacta del modelo de ratón MPTP de la enfermedad de Parkinson. Casi bloquea por completo la tinción de las neuronas TH-positivas para la nitrotirosina, un marcador de daño celular mediado por NO. Este químico (aproximadamente 20 mg/kg/día) atenúa la activación glial inducida por MPTP y previene la pérdida de células dopaminérgicas en la sustancia negra pars compacta (SNpc) en el modelo de ratón MPTP de la enfermedad de Parkinson. Resulta en una reducción del número de microglía activada y astrocitos reactivos en el hipocampo y la corteza de ratones transgénicos APPV717I de 10 meses de edad. El tratamiento con este compuesto reduce la expresión de las enzimas proinflamatorias ciclooxigenasa 2 (COX2) y óxido nítrico sintasa inducible (iNOS). Disminuye los niveles de ARNm y proteínas de la beta-secretasa-1 (BACE1), y también una reducción del 27% en los niveles del péptido Abeta1-42 soluble. |
Referencias |
|
Aplicaciones
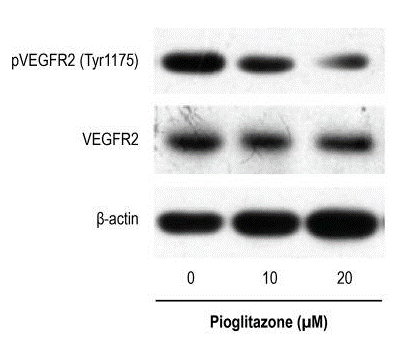
| Métodos | Biomarcadores | Imágenes | PMID |
|---|---|---|---|
| Western blot | p-VEGFR2 / VEGFR Phopsho-RB / RB |
|
29972411 |
Información del ensayo clínico
(datos de https://clinicaltrials.gov, actualizado el 2024-05-22)
| Número NCT | Reclutamiento | Condiciones | Patrocinador/Colaboradores | Fecha de inicio | Fases |
|---|---|---|---|---|---|
| NCT05946564 | Not yet recruiting | ANCA Associated Vasculitis|Rapidly Progressive Glomerulonephritis|Crescentic Glomerulonephritis |
Assistance Publique - Hôpitaux de Paris|Ministry of Health France |
July 2023 | Phase 3 |
| NCT05305287 | Recruiting | Non-Alcoholic Fatty Liver Disease|Type 2 Diabetes|Mitochondrial Metabolism Disorders |
The University of Texas Health Science Center at San Antonio|National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) |
November 1 2022 | Phase 4 |
| NCT05411965 | Completed | Diabetes Mellitus Type 2 |
Boryung Pharmaceutical Co. Ltd |
April 28 2022 | Phase 1 |
| NCT04501406 | Recruiting | Type 2 Diabetes Mellitus (T2DM)|Nonalcoholic Steatohepatitis |
University of Florida|National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) |
December 15 2020 | Phase 2 |
| NCT04535700 | Completed | Type 2 Diabetes |
Fundacion para la Investigacion Biomedica del Hospital Universitario Ramon y Cajal |
September 18 2020 | Phase 4 |
| NCT00904046 | Recruiting | Uric Acid Kidney Stone Disease |
University of Texas Southwestern Medical Center|National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)|Takeda Pharmaceuticals North America Inc. |
September 5 2019 | Not Applicable |
Soporte técnico
Tel: +1-832-582-8158 Ext:3
Si tiene alguna otra consulta, por favor deje un mensaje.
